Section: New Results
Medical Image Analysis
3D/2D Coronary Arteries Registration
Participants : Thomas Benseghir [correspondent] , Grégoire Malandain [Morpheme Team] , Régis Vaillant [GE-Healthcare] , Nicholas Ayache.
This work has been performed in colaboration with GE-Healthcare (Buc) and the Morpheme team at Inria SAM.
3D/2D Registration, Computed Tomography Angiography, X-ray Fluoroscopy, Coronary Arteries, Vascular Tree
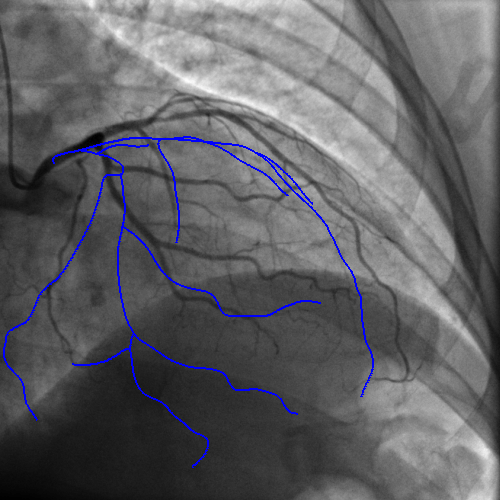
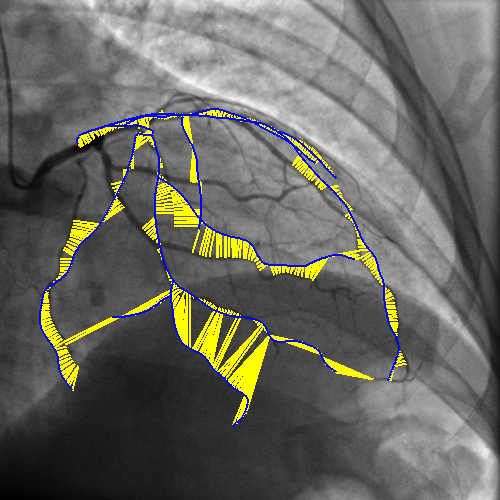
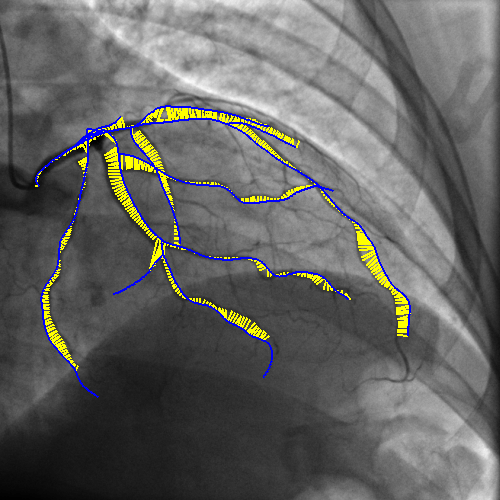
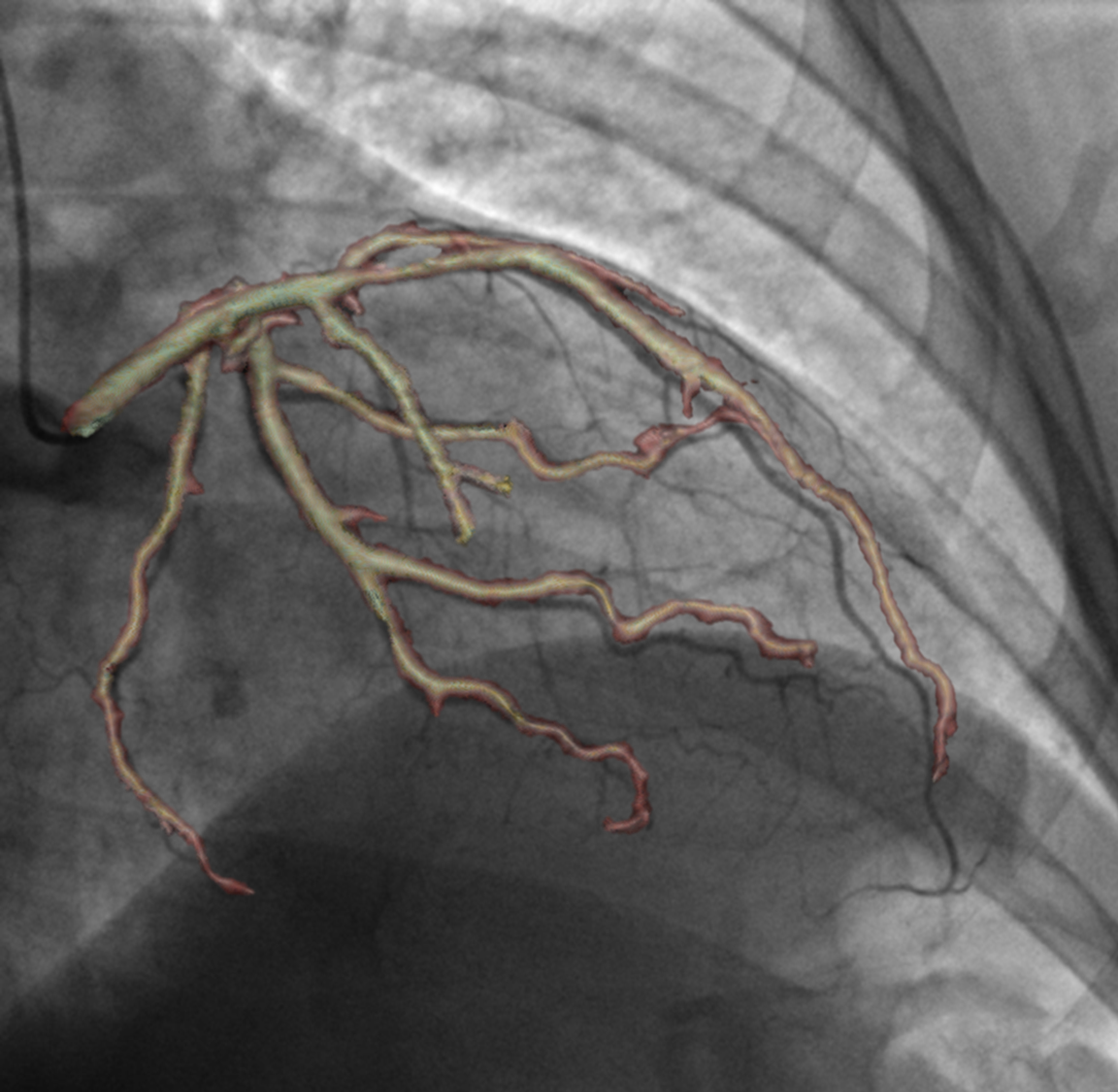
Integrating vessel calcifications and occlusion information, extracted from pre-operative 3D CT angiography images into a live fluoroscopic 2D image can greatly improve the guidance of percutaneous coronary interventions. Such task requires a registration step that must provide relevant correspondences between these two complementary modalities. We are developing a framework aiming at preserving the topology of the vascular structures matched between both images.
The introduction of topology in the pairing procedure allows to decrease the mismatches with respect to geometrically-based pairing procedures (e.g. Iterative Closest Point), which, in turn, improves the success rate of the registration method. This is exemplified by Fig. 3 where the proposed pairing method is compared to ICP.
|
Video Synchronization: An Approach Towards Endoscopic Re-localization
Participants : Anant Suraj Vemuri [correspondent] , Nicholas Ayache.
Endoscopy, Barrett's Esophagus, Re-localization, Electromagnetic Tracking
-
Barrett’s esophagus is the pre-malignant lesion for the majority of patients with esophageal adenocarcinoma. The evolution of the disease involves endoscopic surveillance for patients every 3-6 months, according to the Seattle protocol.
-
The approach is labor-intensive and the primary problem is the inter-operative re-localization of these biopsy sites to guide the treatment.
-
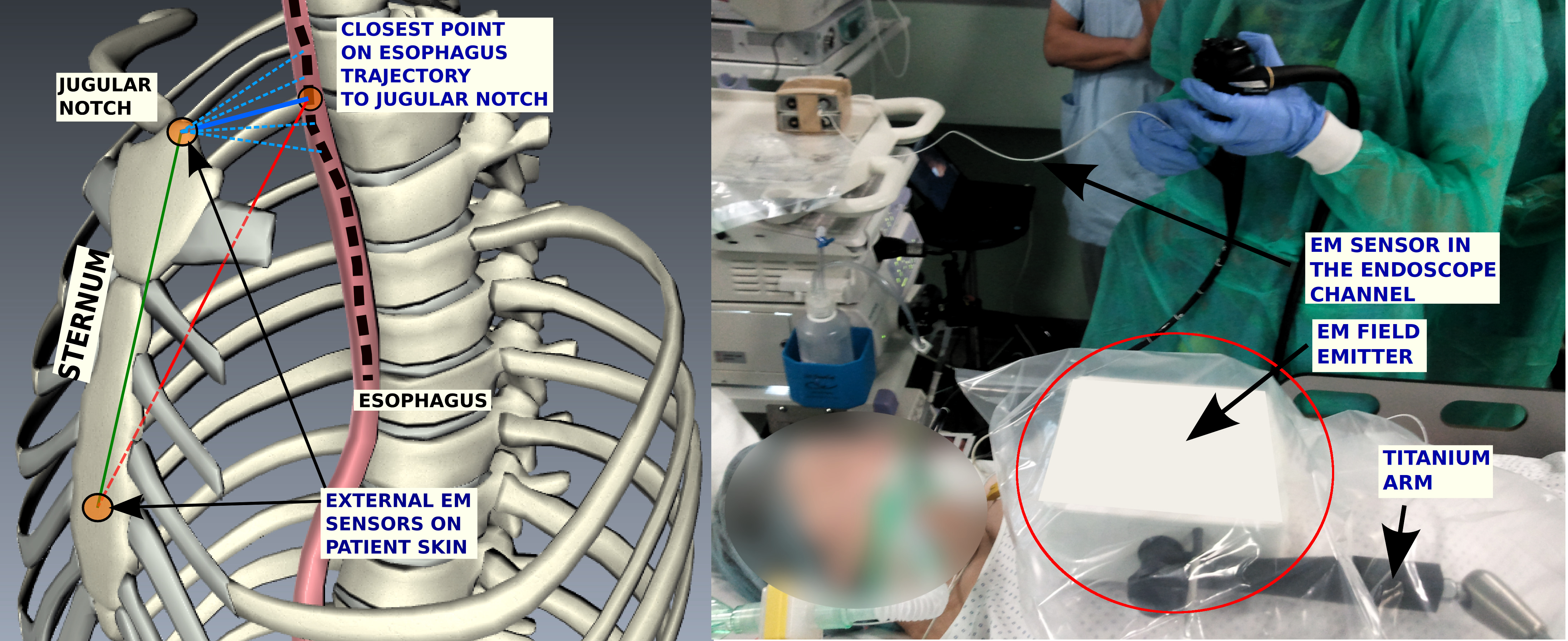
In an earlier work we had proposed a general framework for inter-operative biopsy site re-localization framework by introducing an Electro-magnetic tracking system (EMTS) into the loop and providing a way to inter-operatively register video sequences to provide a guided navigation in the esophagus.
-
This work has been extended further to fit the operating room workflow. Two external landmarks have been added to the system setup as shown in Fig.4 , to obtain a complete reference frame with respect to the patient and to make the registration, patient specific [38] . The patient-localized reference frame allows the recovery of complete including the roll angle about the esophageal axis.
|
A sparse Bayesian framework for non-rigid registration
Participants : Loic Le Folgoc [correspondent] , Hervé Delingette, Antonio Criminisi, Nicholas Ayache.
This work has been partly supported by Microsoft Research - Inria joint laboratory through its PhD Scholarship Programme and the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Registration, Automatic Relevance Determination, Uncertainty Quantification
We propose a sparse Bayesian framework for non-rigid registration. It provides a principled approach to efficiently find an optimal, sparse parameterization of deformations among any preset, widely overcomplete range of basis functions. It addresses open challenges in state-of-the-art registration, such as the automatic joint estimate of model parameters (e.g. noise and regularization levels). We have evaluated the feasibility and performance of our approach on cine MR, tagged MR and 3D US cardiac images, and show state-of-the-art results on benchmark datasets evaluating accuracy of motion and strain (see Fig.5 ). This work was presented during the MICCAI 2014 conference[20] .
|
Segmentation and anatomic variability of the cochlea and other temporal bone structures from medical images
Participants : Thomas Demarcy [correspondent] , Hervé Delingette, Clair Vandersteen, Dan Gnansia [Oticon Medical] , Nicholas Ayache.
This work is funded by a CIFRE grant involving Oticon Medical (Vallauris) and performed in collaboration with the IUFC Nice (Pr. Guevara) and CHU Nice (Pr. Raffaelli) .
image segmentation ; surgery planning ; shape modelling ; anatomic variability ; cochlear implant ; temporal bone
-
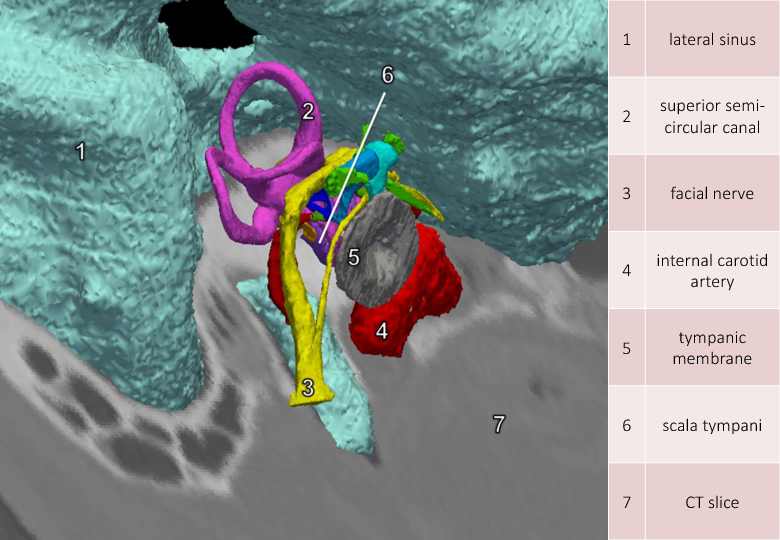
We applied semi-automatic segmentation methods to extract anatomical structures on the inner ear on both micro-CT and CT scan images.
-
-CT and CT images acquired on the same subject were fused with their segmentation.
-
We designed a teaching tool[37] for advanced visualization of temporal bone structures (see Fig. 6 ).
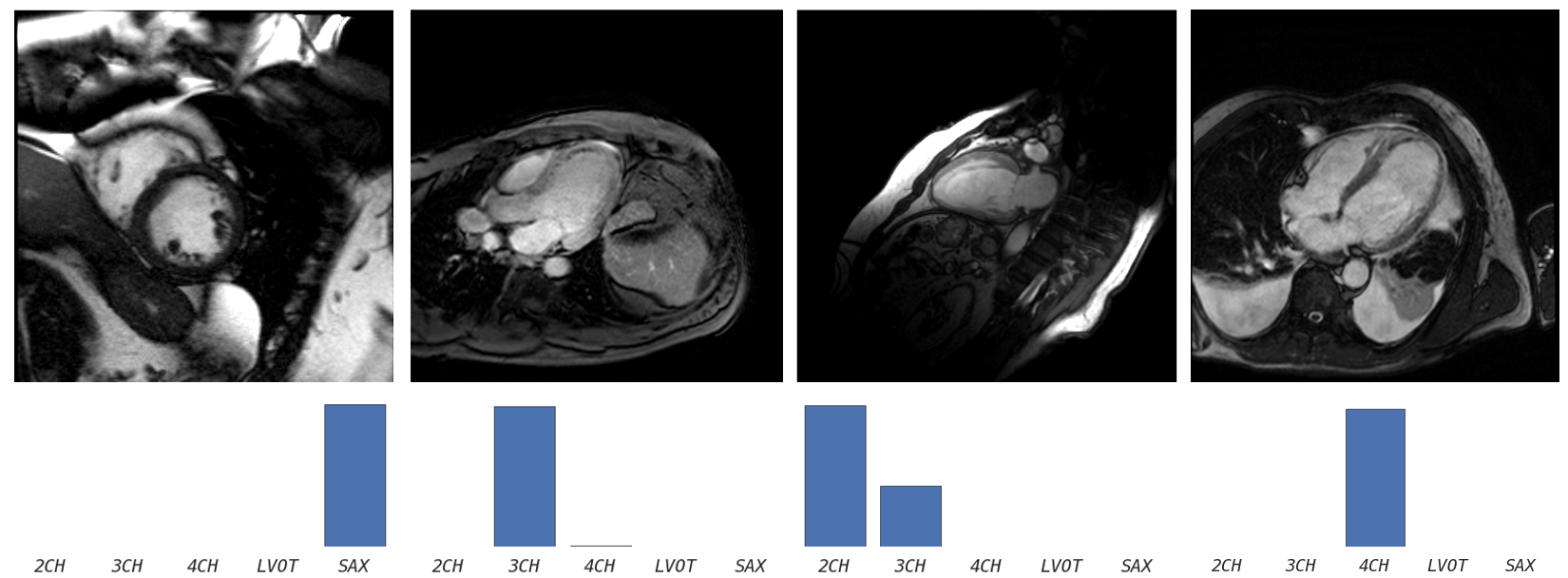
Understanding cardiac planes of acquisition
Participants : Jan Margeta [correspondent] , Nicholas Ayache, Daniel C Lee [Northwestern University] , Antonio Criminisi [Microsoft Research Cambridge] .
This work has been partly supported by Microsoft Research through its PhD Scholarship Programme, by ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images), and by the VP2HF FP7 research project.
Cardiac imaging, Machine learning, Magnetic resonance, Data wrangling
DICOM image format defines several tags by which the images can be queried and filtered. Many useful tags are however not standardized and must be cleaned prior to any large scale analysis.